Specifically remove oligosaccharides from proteins
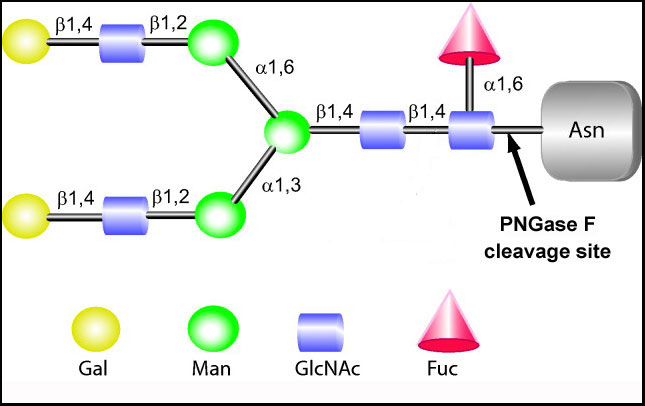
PNGase F PRIME and PNGase F PRIME-LY (lyophilized version) are a mutant recombinant PNGase F cloned from Flavobacterium meningosepticum and expressed and purified from E. coli. The proprietary changes made to PNGase F have been shown to have unique characteristics when compared to other commercially-available sources of PNGase F. Data generated by independent labs shows that PRIME works on native glycoproteins and serum glycoproteins in minutes at room temperature. Glycan analysis of the digestion products shows that PNGase F PRIME digestion led to more complete glycan release and also allowed for the cleavage of glycans not released by the commercially-available enzymes when used at the same concentrations with the same digestion conditions. This advancement benefits applications that seek to understand glycobiology in a natural milieu. Preliminary data indicates that PNGase F PRIME has a higher specificity towards complex (tri and tetra-antennary) sialylated structures compared to the commercially sourced enzyme. Additionally, the work presented in this Analytical Chemistry paper utilized PNGase F PRIME for all in situ tissue work as the commercially-available PNGase enzymes did not work on native tissue to allow glycan recognition.
PNGase PRIME-LY ULTRA Glycosidases are a low salt version which is specifically optimized for fine structure Mass Spec Imaging where high resolution imaging in small structures or cells is needed. This high-performance enzyme has been shown to require as few as 3,000 cells per replicate with 3-20% coefficient of variation to capture label-free N-glycan profiles (as shown in Angel et. al., J Proteome Res. 2019).
- Excellent for use in high-end use applications especially those using or requiring UPLC/HPLC, Hydrophilic Interaction Chromatography (HILIC), and/or MALDI-Glycan Mass Spec Imaging.
- Released glycans can be examined following labeling with the Waters RapiFluor-MS dye and analyzed by normal phase hydrophilic interaction chromatography (HILIC) using various HPLC/UPLC-based systems.
- Imaging of released glycans directly on tissue can be done following spraying of enzyme on tissue and incubation at 37°C for 1 hour. (At our standard concentration of 2mg/mL, one 50 µL vial can make 4 slides.) Glycan can be detected using instruments such as the Bruker Daltonics SolariX™ 7T Hybrid FTMS System, a Bruker Daltonics RapifleXTM MALDI Tissuetyper, or a Bruker Daltonics UltrafleXtreme MALDI-TOF/TOF mass spectrometer.
Peak-a-boo we see you
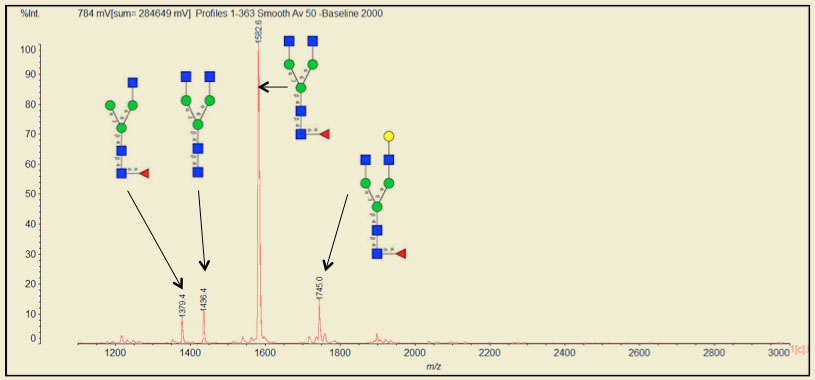
Analysis by mass spectrometry is when PRIME truly shines. Traditional PNGase provides fewer and blunted peaks as compared with PRIME. In nearly every side-by-side study to date, PNGase F PRIME produces favorable results. For example, native Avastin® (IgG1 based therapeutic for cancers) treated with PRIME releases 11 different oligosaccharides structures (only a subset of data shown below).
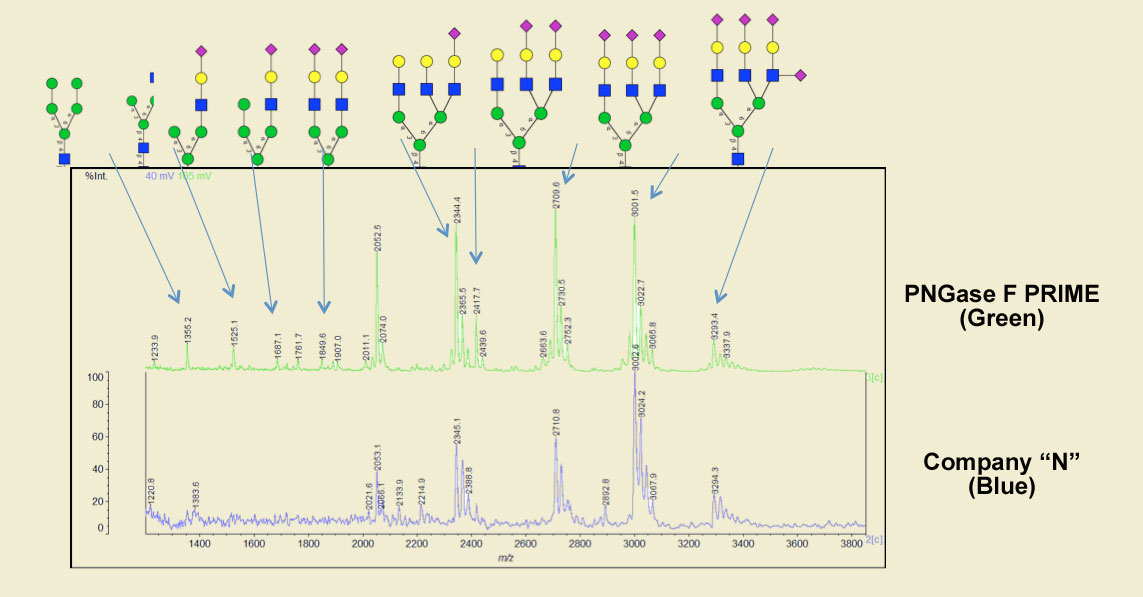
In another example, when compared with a very popular PNGase F, PRIME produced 33% more peaks, sharper peaks and with much lower background when cleaving the native form of the blood borne glycoprotein fetuin.
The same treatments show marked differences even by HPLC analysis. PNGase F PRIME (in blue) shows more pronounced peaks than the same competing PNGase F (in black). Also note the additional peaks in both the neutral and sialylated regions, particularly the trisialo region.
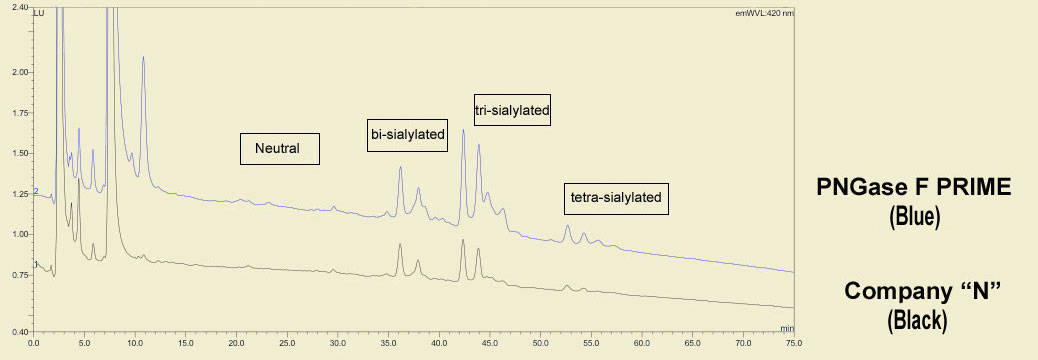
This type of richer data presents the opportunity to develop a deeper, more complete, understanding of glycobiology.
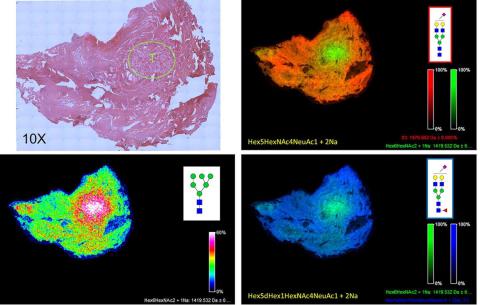
N-glycan Imaging of a Prostate Cancer FFPE Tissue
A 5 micron FFPE slice of a human prostate cancer (Gleason 7) was antigen retrieved and incubated with PNGase F PRIME™ as described in Powers et al. Images were acquired using a 7T MALDI-FTICR Solarix mass spectrometer and FlexImaging 4.1 software. Shown in the upper right panel is a H&E stained image of the tissue (at 10X magnification), with the area of the highest density of tumor shown in the circle, marked with a T. The bottom left panel shows a typical tumor N-glycan pattern for a single glycan, an Man6 at m/z=1419. In the top right panel and bottom right panel, a two-glycan overlay is shown for the tumor glycan at m/z = 1419 (in green) and a mono-sialylated biantennary glycan at m/z = 1976 (top right panel, orange) and a mono-sialylated and core fucosylated glycan at m/z = 2122 (bottom right panel, blue).
The following citations provide much greater detail of this research that utilized PRIME™:
Powers TW, Neely BA, Shao Y, Tang H, Troyer DA, Mehta AS, Haab BB, Drake RR. (2014) MALDI Imaging Mass Spectrometry Profiling of N-Glycans in Formalin-Fixed Paraffin Embedded Clinical Tissue Blocks and Tissue Microarrays. PLoS One. 9(9):e106255
Powers, T.W. Holst, S., Wuhrer, M.,Mehta, A.S., Drake, R.R. (2015) Two-Dimensional N-Glycan Distribution Mapping of Hepatocellular Carcinoma Tissues by MALDI-Imaging Mass Spectrometry. Biomolecules, 5, 2554-2572.
The following citations provide much greater detail of this research that utilized PRIME™ ULTRA
Angel P. M., Saunders J., Clift C. L., White-Gilbertson S., Voelkel-Johnson C., Yeh E., Mehta A., Drake R. R. (2019) A Rapid Array-based Approach to N-glycan Profiling of Cultured Cells. J. Proteome Res. 2019, 18, 3630–3639.
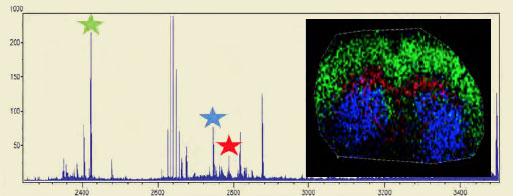
MALDI-IMS Analysis of a Mouse Brain Slice
The three starred oligosaccharide groups in the mass spec trace to the right are color coded and overlayed on a mouse brain section. The distribution shows a distinct localization pattern.
PNGase F Prime Glycosidase - Lyophillized

NZPP010LY – PNGase F PRIME-LY, 1 x vial w/100ug lyophilized (50,000 units total)
$155.00 Add to cartPNGase F Prime Glycosidase - Lyophillized ULTRA
PNGase F Prime Glycosidase
|
SPECIFICATIONS
|
|
|---|---|
| Part Numbers | NZPP010, NZPP010LY, NZPP050LY, NZPP550LY, NZPULT10LY |
| Target | Asparagine-linked (N-linked) oligosaccharides (mannose, hybrid, and complex) |
| Compatible Glycoprotein | Native and denatured |
| Source of PNGase F Prime | Recombinant, E. coli |
| Total Protocol Length | 30 minutes to 24 hours |
| Typical Yield | 1 Unit of enzyme will completely deglycosylate 10 µg of denatured RNase B following incubation for 10 minutes at 37°C. 1 Unit = 1 IUB milliunit. |
| Concentration | PNGase F Prime – 1,000,000 units/ml (2mg/mL)
PNGase F Prime-LY – 100ug per vial, lyophilized from 2mg/mL stock solution. PNGase F Prime-LY ULTRA – 100ug per vial, lyophilized from low salt stock solution. Low salt optimized for fine structure Mass Spec Imaging. |
| Purity | ≥95% as determined by SDS PAGE |
| Contains Glycerol | No |
| Compatible with Mass Spec | Yes |
| Affinity tag for purifcation | Yes. PNGase F Prime contains a His-tag. |
| Reaction Conditions | In vitro and in situ |
| Storage | -20°C or 4°C (avoid multiple freeze-thaws) |
| Shelf Life | 1 year |
| Microbe Free | Yes, lot tested |